Abstract
Electroencephalography (EEG) measures electrical brain activity and is a common tool for monitoring neurodevelopment and diagnosing brain disorders. However, analysing EEG data is challenging due to its complex interpretability and inter-subject variability. Traditional techniques often rely on handcrafted feature engineering and do not scale or generalise well. Deep learning models present a promising solution to these issues, as they have already demonstrated success in capturing long-term dependencies and modelling sequential data, such as time series and language modelling. For this reason, we adapt a pre-trained Transformer- and State-space-based model for processing a longitudinal EEG dataset from South African infants, for which there is limited literature and evidence of deep learning applications in this subject domain. We aim to determine whether current state-of-the-art EEG transformers can handle this type of data and potentially predict neurodevelopmental delays, a key research area for understanding infant brain maturation. We investigate this by performing Bayley scale score prediction (a scoring system used as a developmental assessment) on both a model trained from scratch and a pretrained model. Our results indicate that starting with a pre-trained model is favourable, but overall performance remains limited unless deep learning models have access to much larger infant EEG datasets. This study provides an initial benchmark and clarifies the current limitations of transformer and state-space models for brain developmental research using infant EEG.
Videos 1
Watch presentations, demos, and related content
Demonstration video on the deepEEG project: Deep Learning for Infant-Brain EEG Modelling
Like, comment, and subscribe on YouTube to support the creator!
Demonstration video on the deepEEG project: Deep Learning for Infant-Brain EEG Modelling
Gallery 6
Explore the visual story of this exhibit

deepEEG
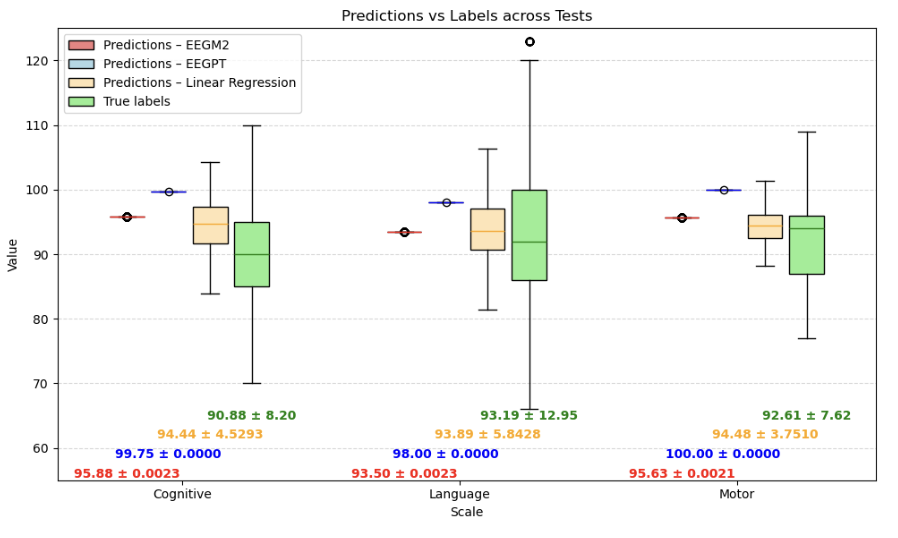
Bayley scale true scores vs. predictions across deep learning models and baselines
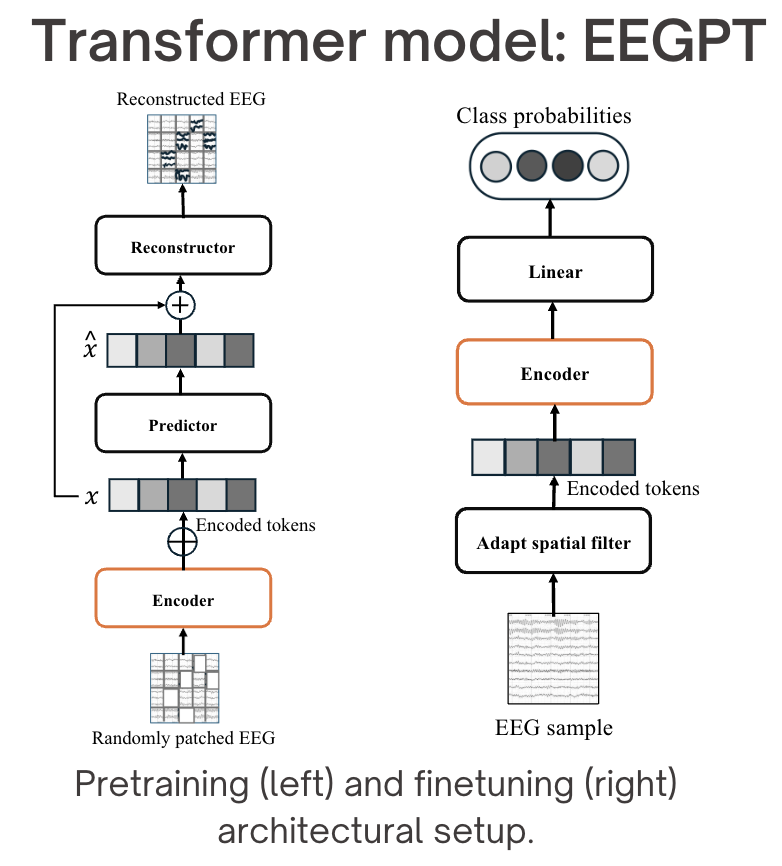
Architecture overview of EEGPT
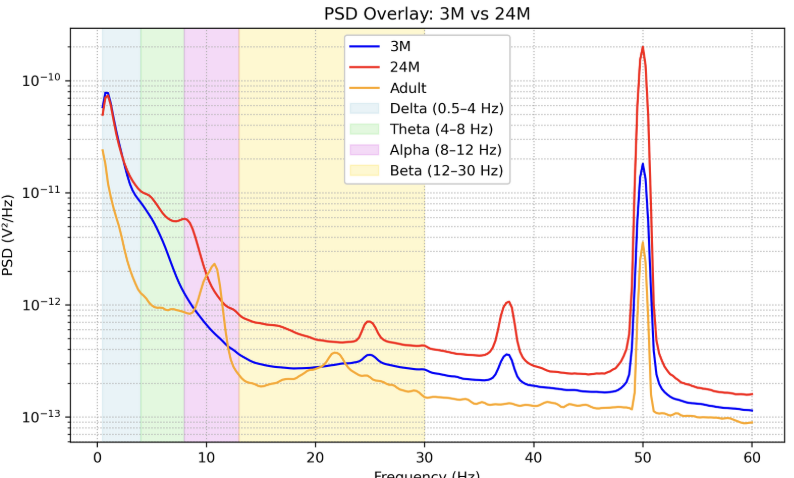
Power spectral density plot of the EEG samples from the Khula dataset
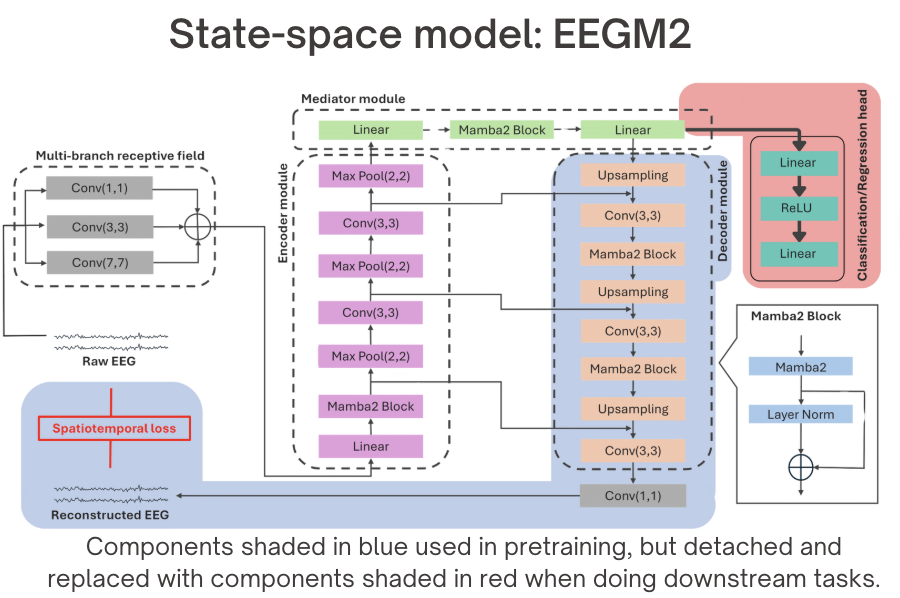
Architecture overview of EEGM2
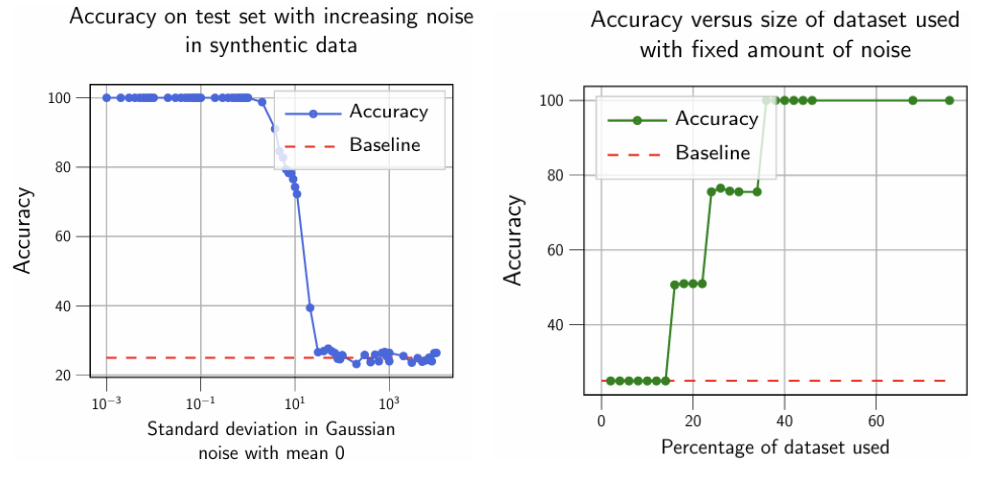
Synthetic data experiment results