ClusterMol
Exploring clustering techniques to find dominant conformations of flexible carbohydrate molecules.
Talk to us
No meetings are currently scheduled.
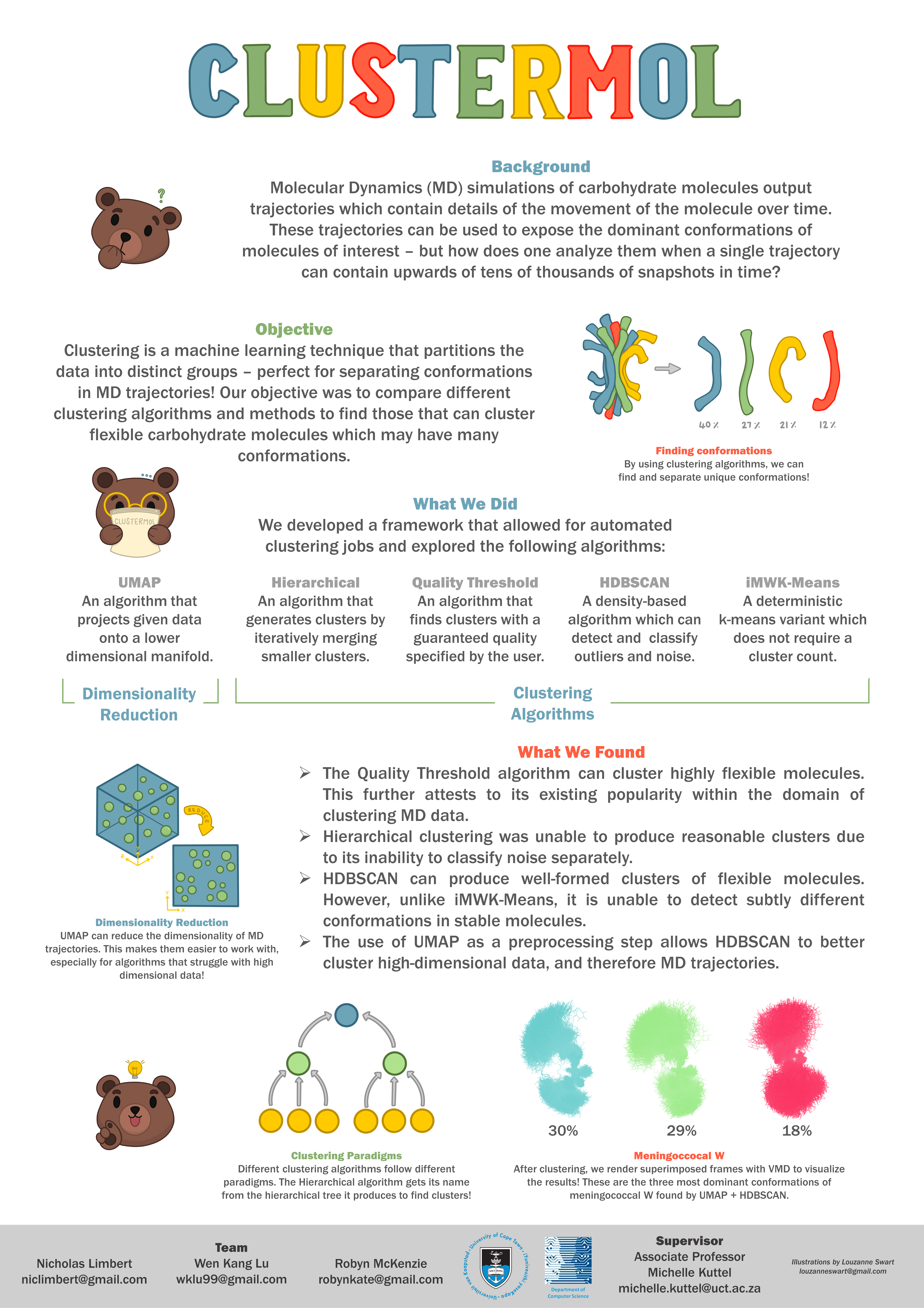
Molecular Dynamics (MD) simulates the behaviour of molecules which are allowed to interact with one another over a fixed period of time; it outputs what is known as a trajectory of the behaviour observed. This trajectory is a record of the movement of every atom during the simulation, with respective changes in position over time calculated by solving Newton’s equations of motion. Since the effects of molecular interactions occur so rapidly, positions are often updated at the femtosecond timescale. Therefore, trajectories containing even just a few nanoseconds worth of time frames amount to millions of locational data points to observe.
MD simulations play an important role for researchers interested in the effects molecular conformations can have in their applications, some of which include drug-receptor interaction and protein folding. Consequently, it is imperative that effective methods exist to gather insight from trajectories as efficiently as possible. A common way to do so is through the use of clustering algorithms that can partition the trajectory into a set of dominant conformations that are deemed structurally unique from one another. This project explored different clustering techniques for clustering flexible carbohydrate molecules like polysaccharides.
Images